CheShift
Validate your protein model with chemical shift predictions
What is CheShift? Complete Overview
CheShift is a specialized tool designed for researchers and scientists working with protein models. It provides theoretical chemical shift predictions (13C and 13N) and validation capabilities to identify potential flaws in protein structures. The tool helps biochemists, structural biologists, and computational chemists improve the accuracy of their protein models by comparing theoretical predictions with experimental data. CheShift addresses the critical need for reliable validation methods in protein modeling, offering both chemical shift calculations and structural validation features. The tool is particularly valuable for academic researchers, pharmaceutical scientists, and anyone working with protein structure prediction and validation.
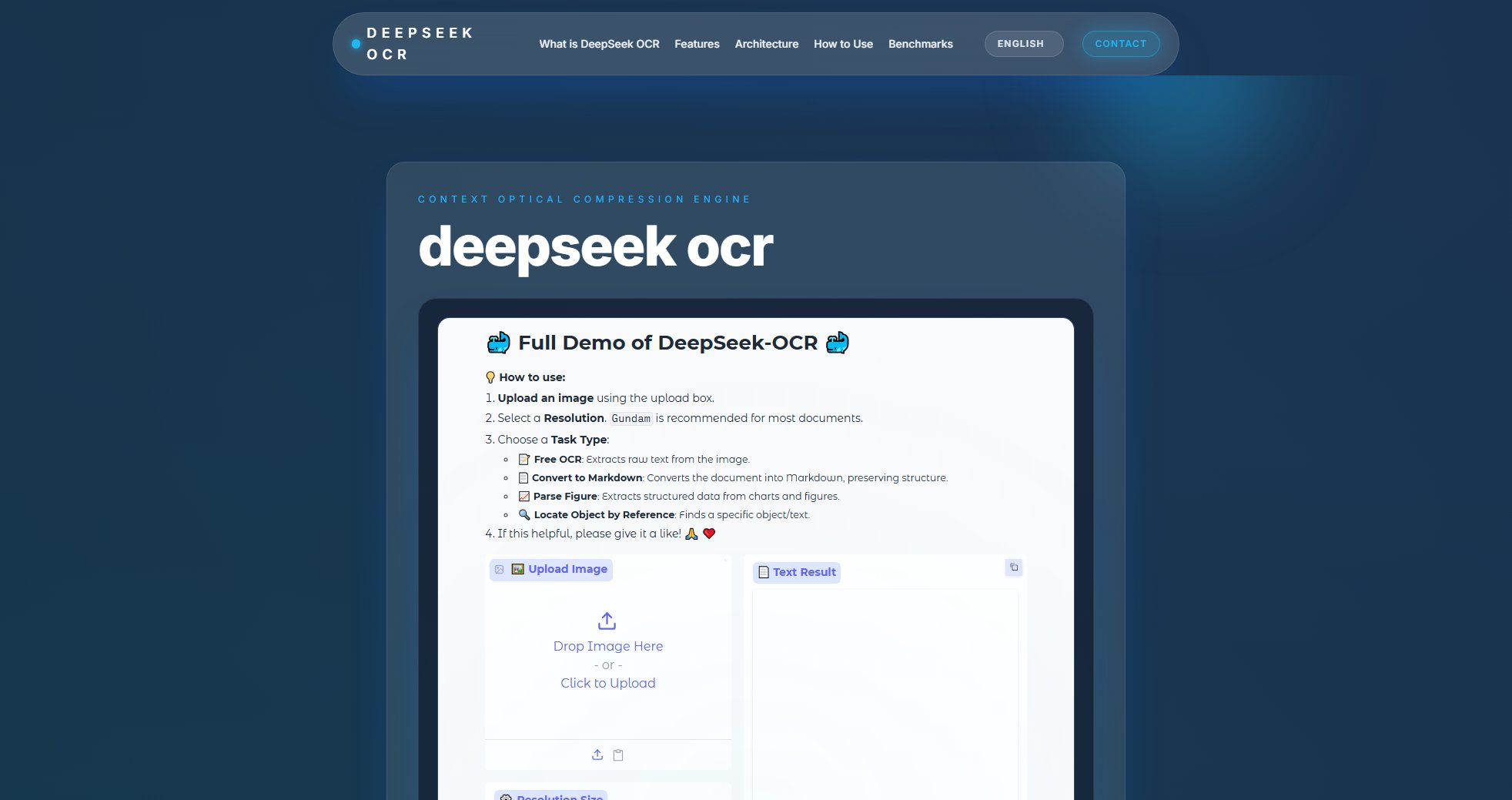
CheShift Interface & Screenshots
CheShift Official screenshot of the tool interface
What Can CheShift Do? Key Features
13C and 13N Chemical Shift Prediction
CheShift calculates theoretical chemical shifts for carbon-13 (13C) and nitrogen-13 (13N) atoms in protein models. This feature provides researchers with a comprehensive list of predicted chemical shift values that can be compared with experimental NMR data. The predictions help in assessing the quality and accuracy of protein structural models.
Protein Model Validation
The tool offers advanced validation capabilities by comparing theoretical chemical shifts with experimental data. It generates a colored 3D protein model visualization that highlights potential structural flaws, making it easier for researchers to identify problematic areas in their protein structures.
Structural Solution Suggestions
Beyond identifying flaws, CheShift provides a list of potential solutions for some of the detected issues in protein models. This proactive feature helps researchers quickly address structural problems and improve their models' accuracy.
PyMOL Integration
CheShift offers a plugin for PyMOL, the popular molecular visualization system. This integration allows users to seamlessly incorporate chemical shift validation into their existing PyMOL workflows, enhancing their structural analysis capabilities.
Visual Flaw Representation
The tool generates color-coded 3D visualizations of protein models where potential structural problems are clearly highlighted. This intuitive representation helps researchers quickly grasp where their models might need improvement.
Best CheShift Use Cases & Applications
Protein Structure Refinement
Researchers can use CheShift to validate newly predicted protein structures against experimental NMR data, identifying areas that may need refinement to better match the experimental observations.
Model Quality Assessment
Before publishing or using protein models for drug discovery, scientists can employ CheShift to assess model quality, increasing confidence in their structural predictions.
Teaching and Education
Instructors can use CheShift in biochemistry or structural biology courses to demonstrate the relationship between protein structure and NMR chemical shifts, providing students with practical validation tools.
Drug Discovery Research
Pharmaceutical researchers can validate protein-ligand complex structures, ensuring accurate representation of binding sites for more reliable drug design and discovery processes.
How to Use CheShift: Step-by-Step Guide
Prepare your protein model in a compatible format (typically PDB format) that you want to analyze or validate using CheShift.
If using the PyMOL plugin, load your protein model in PyMOL and activate the CheShift plugin. For standalone use, upload your model file to the CheShift interface.
Select the type of analysis you want to perform: either chemical shift prediction or model validation using existing chemical shift data.
Run the analysis. For chemical shift prediction, CheShift will calculate and return theoretical values. For validation, it will compare your model with experimental data.
Review the results: examine the list of predicted chemical shifts or the color-coded 3D model showing potential structural flaws along with suggested solutions.
CheShift Pros and Cons: Honest Review
Pros
Considerations
Is CheShift Worth It? FAQ & Reviews
CheShift primarily works with protein models in PDB (Protein Data Bank) format, which is the standard format for protein structure representation.
Currently, CheShift specializes in predicting chemical shifts for carbon-13 (13C) and nitrogen-13 (13N) atoms in protein structures.
No advanced programming knowledge is needed for basic use. The PyMOL plugin provides a graphical interface, and the standalone version typically requires only file uploads.
While predictions are theoretical, they are based on established algorithms and can provide valuable insights when compared with experimental data for validation purposes.
CheShift is primarily designed for protein structures and may not provide optimal results for nucleic acids or other biomolecules.